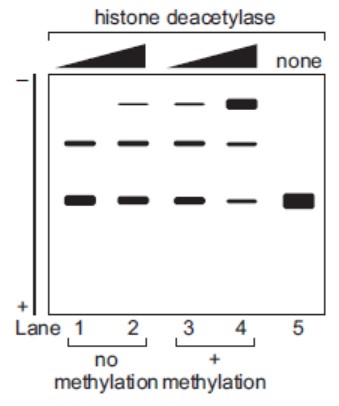
#Question id: 3706
#Unit 3. Fundamental Processes
During the elongation stage of DNA replication
#Question id: 3707
#Unit 3. Fundamental Processes
The replication of DNA is ________ because ________.
#Question id: 3708
#Unit 3. Fundamental Processes
The incorporation of new nucleotides into a growing strand of DNA occurs during ________.
#Question id: 3709
#Unit 3. Fundamental Processes
DNA that has been liberally labeled with radioactive 14C is used as the template for replication. Replication is carried out in a medium containing only unlabeled nucleotides. After two rounds of replication, what percent of DNA molecules are still labeled?
#Question id: 3710
#Unit 3. Fundamental Processes
The protein machine that carries out DNA replication is called ________.
#Question id: 3711
#Unit 3. Fundamental Processes
How many replisomes are required for one complete round of DNA replication in E. coli?