#Question id: 2837
#Unit 2. Cellular Organization
#Question id: 2838
#Unit 2. Cellular Organization
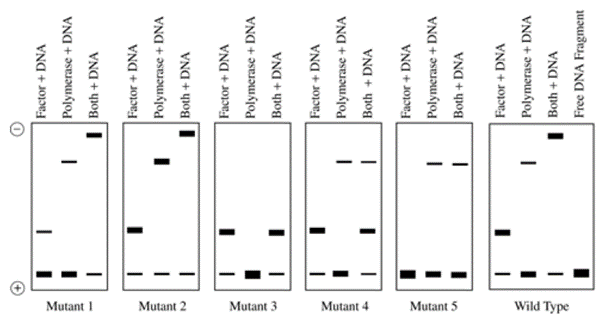
You want to study the potential interaction between nucleosome-bound DNA and a specific histone deacetylase. You decide to perform an electrophoretic mobility shift assay (EMSA). You use a 32P end-labelled, linear template DNA that contains two nucleosome positioning sites. You assemble two nucleosomes on the DNA template before incubation with and without the histone deacetylase. For some reactions, you use unmodified nucleosomes. For other reactions, you use nucleosomes that are methylated at lysine 36 of the histone protein H3.
A. The histone deacetylase binds nucleosome bound-DNA in lanes 1, 2, 3, and 4.
B. The histone deacetylase binds nucleosome bound-DNA in lanes 3& 4.
C. The histone deacetylase seems to recognize methylated nucleosomes at lysine 36 of histone H3 in lane 1, 2 & 3 better than unmethylated nucleosomes in lane 4 &5
D. The histone deacetylase seems to recognize methylated nucleosomes at lysine 36 of histone H3 in lane 1 & 2 better than unmethylated nucleosomes in lane 3 &4
#Question id: 2839
#Unit 2. Cellular Organization
Consider for correct match
|
I. All the different members of family of genes and proteins are sufficiently similar in sequence to suggest a common ancestral sequence. |
A. Homologous |
|
II. Sequences that presumably diverged as a result of gene duplication |
B. Paralogous |
|
III. Sequences that arose because of speciation |
C. Orthologous |
#Question id: 2840
#Unit 2. Cellular Organization
An experimental setup provided the assay for identifying factors that facilitate transcription in the presence of chromatin. A factor called FACT (facilitates chromatin transcription) functions as
#Question id: 2841
#Unit 2. Cellular Organization
To duplicate a chromosome, at least half of the nucleosomes on the daughter chromosomes must be newly synthesized. But histone proteins as new and old occupies strands differently as
#Question id: 2842
#Unit 2. Cellular Organization
Epigentic code for inactive X chromosome have post-translational modifications characteristic of other regions of heterochromatin as